Dec 2, 2008
Interview questions - phd or post doc
1. What characteristics distinguish this program from others in the same field ?
2. Tell me abt yourself ? 3. What are your strengths and weakness ? 4. If your are not accepted into graduate school ? What are your plans ?
5. Why did you choose this career ?
6. What do you know about our program ? / Why did you choose to apply to our program ? 7. What do you believe your greatest challenge will be if you are accepted into this program ?
8. In college, what courses did you enjoy the most? The least ? Why?
9. How would your professors describe you?
10. How will you be able to make a contribution to this field?
11. What are your hobbies?
12. Explain a situation in which you had a conflic and how you resolved it. What would you do differently? Why? 13. Describe your greatest accomplishment?
14. Tell me about your experience in this field? What was challenging? What was your contribution?
15. What are your career goals? How will this program help you achieve your goals ? 16. What skills do you bring to the program. How will you help your mentor in his or her research ? / Why should we take you and not someone else? / In what ways have your previous experience prepared you for graduate study in our program ?
17. Are you motivated? Explain and provide examples. 18. What do you plan to specialize in ? 19. What can be determined about an applicant at an interviews?
20. Has anything ever irritated you about people you have worked with?
21. Is there anyone you just could not work with?
22. Would you rather be liked or feared ?
23. What has been the biggest obstacle you have overcome? 24. By what process do you make decisions?
25. What do you do when you are right and others disagree with you? 26. How would you supervise a postgraduate researcher?
27. Who are the key researchers in your field? 28. What are the most important rewards you expect in your career? 29. how do you determine or evaluate success?
28. How do you want to see you in next 10 years?
29. Basic scientific questions in your area?
Oct 29, 2008
Prediction of Protein Structure In 1D - Secondary Structure, Membrane Regions, and Solvent Accessibility
Burkhard Rost
Structural Bioinformatics, P Bourne & H Weissig (eds.), Wiley, 2007, in press
http://cubic.bioc.columbia.edu/papers/2008_rev_1dpred/paper.html
Free English/metric conversion software
Oct 18, 2008
Free education online
Oct 16, 2008
Free online MIT Courses
Nobel Week
Every year since 1901 the Nobel Prize has been awarded for achievements in physics, chemistry, physiology or medicine, literature and for peace. The Nobel Prize is an international award administered by the Nobel Foundation in Stockholm, Sweden. In 1968, Sveriges Riksbank established The Sveriges Riksbank Prize in Economic Sciences in Memory of Alfred Nobel, founder of the Nobel Prize. Each prize consists of a medal, personal diploma, and a cash award
Why is the Nobel Peace Prize awarded in Oslo and all the other Nobel Prizes in Stockholm?
Alfred Nobel left no explanation as to why the prize for peace was to be awarded by a Norwegian committee while the other four prizes were to be handled by Swedish committees. In the will he wrote:
"The prizes for physics and chemistry shall be awarded by the Swedish Academy of Sciences; that for physiology or medical works by the Karolinska Institute in Stockholm; that for literature by the Academy in Stockholm, and that for champions of peace by a committee of five persons to be elected by the Norwegian Storting."
Why do you use the word Nobel Laureate and not Nobel Prize Winner?
The awarding of the Nobel Prizes is not a competition or lottery, and therefore there are no winners or losers. Nobel Laureates receive the Nobel Prize in recognition of their achievements in physics, chemistry, physiology or medicine, literature, or peace.
Nobel Prize amount
10000000(sweden kronor) = 6,71,54,875 (Indian rupees) [in 2007]
Nobel Laureates Facts (2007)
807 Nobel Laureates!
787 individuals and 20 organizations have been awarded the Nobel Prize. Some Laureates and organizations have been awarded more than once.
Only 35 women!
35 Nobel Laureates have been women, and the remaining 752 were men.
The youngest Nobel Laureate
To date, the youngest Laureate is Lawrence Bragg, who was just 25 years old when he received the Nobel Prize in Physics with his father in 1915.
The oldest Nobel Laureate
The oldest Laureate to date is Leonid Hurwicz, 2007 Economics Laureate, who is 90 years old.
Which person or organization has received the Nobel Prize more than once?
Marie Curie received the 1903 Nobel Prize in Physics, and the 1911 Nobel Prize in Chemistry;
John Bardeen received the 1956 and the 1972 Nobel Prize in Physics;
Frederick Sanger received the 1958 and the 1980 Nobel Prize in Chemistry;
Comité International de la Croix Rouge received the 1917, 1944 and 1963 Nobel Peace Prize;
Office of the United Nations High Commissioner for Refugees received the 1954 and the 1981 Nobel Peace Prize;
Linus Pauling received the 1954 Nobel Prize in Chemistry and the 1962 Nobel Peace Prize.
Who is the only person to have been awarded two unshared Nobel Prizes?
Linus Pauling was awarded the 1954 Nobel Prize in Chemistry and the 1962 Nobel Peace Prize.
The Nobel Peace Prize 2008
conflicts"
Name : Martti Ahtisaari
Country: Finland

The Nobel Prize in Literature 2008
Country : France
"author of new departures, poetic adventure and sensual ecstasy, explorer of a humanity beyond and below the reigning civilization"
 |
The Nobel Prize in Physiology or Medicine 2008
 |  |  |
| Harald zur Hausen | Françoise Barré-Sinoussi | Luc Montagnier |
|---|---|---|
| Germany | France | France |
| German Cancer Research Centre Heidelberg, Germany | Regulation of Retroviral Infections Unit, Virology Department, Institut Pasteur Paris, France | World Foundation for AIDS Research and Prevention Paris, France |
The Nobel Prize in Chemistry 2008
 |  |  |
| Osamu Shimomura | Martin Chalfie | Roger Y. Tsien |
|---|---|---|
| USA | USA | USA |
| Marine Biological Laboratory (MBL) Woods Hole, MA, USA | Columbia University New York, NY, USA | University of California San Diego, CA, USA |
The Nobel Prize in Physics 2008
 |  |  |
| Yoichiro Nambu | Makoto Kobayashi | Toshihide Maskawa |
|---|---|---|
| USA | Japan | Japan |
| Enrico Fermi Institute, University of Chicago Chicago, IL, USA | High Energy Accelerator Research Organization (KEK) Tsukuba, Japan | Kyoto Sangyo University; Yukawa Institute for Theoretical Physics (YITP), Kyoto University Kyoto, Japan |
Nobel prize in Economics 2008
 |
| Princeton University USA |
Reference : http://nobelprize.org/index.
Oct 15, 2008
Oct 3, 2008
Proteopedia
Sep 24, 2008
Web link reference manager
Useful one check it out this http://www.connotea.org/ and sign up (similar to mail sign up) and make use of it.
Sep 23, 2008
Vadlo - Search engine for research scholar
Sep 12, 2008
Protein crystallography
Check it out this site, if you are interested in protein crystallography http://proteincrystallography.org/
Aug 30, 2008
AIGore Nobel Lecture
http://nobelprize.org/nobel_prizes/peace/laureates/2007/gore-lecture_en.html
Aug 26, 2008
The softer side of science
Mastering soft skills helps master one's career.
Scientists should shield themselves from discouraging events and develop a 'frustration tolerance' for paper and grant rejections, criticism by well-meaning colleagues and the depressing tedium of data collection. Then there's hypermotivation. To avoid burn-out, try relaxation (sports, yoga, meditation) and a healthy social life. Just as learning requires a quiet consolidation phase to store material in long-term memory, success in science needs intermittent silence.
Personality traits cannot usually be changed, but there are ways to improve one's disposition in the lab. Perfection, for example, can only be expected in pure mathematics or fairy tales. Beware of the 80–20 rule: for perfection, the last 20% of a task may take 80% of the effort — not a wise choice if you want to be productive. Worse, perfectionism is a sure path to leaving projects or papers incomplete.
A related trap is failure to bring a project to publication. A finished experiment may satisfy your curiosity, but data are only of value to your CV and the rest of the world if published. The drive to completion is healthy, if you wish to succeed in science. Many of us have papers that are 90% finished but never submitted. Consider the time you have already invested, how little is left to do and how much effort it would take to get to the same point with another project.
Writer's block can be a major challenge: sitting in front of a blank page, lacking the wherewithal to start writing. This may be linked to the perfectionist trap ("I do not write unless my text is perfect"), but it could also be the result of a lack of ideas, of writing ability or of self-confidence. It is usually the first 10 minutes of writing that are hardest. Try to start writing without worrying about the presentation or structure.
Scientific leadership qualities — exhibiting responsibility, flexibility and trust — are also essential in all aspects of science.
These seemingly simple tasks can be taxing. But improving soft skills is a critical element of science success.
From Nature magazine http://www.nature.com/naturejobs/2008/080529/full/nj7195-694b.html#top
Jul 24, 2008
retracted paper from india
I have gone through this paper recently which is retracted from India
RETRACTED: A computational docking study for prediction of binding mode of diospyrin and derivatives: Inhibitors of human and leishmanial DNA topoisomerase-I
Bioorganic & Medicinal Chemistry Letters, Volume 17, Issue 18, 15 September 2007, Page 5281
Sandeep Chhabra, Pooja Sharma, Nanda Ghoshal.
Reference : Bioorganic & Medicinal Chemistry Letters
Jun 7, 2008
Gromacs-tutorial
http://md.chem.rug.nl/education/mdcourse/MDpract.html
May 22, 2008
Research article with reviewers comment in Biologydirect
Various hypotheses on MHC evolution suggested by the concerted evolution of CD94L and MHC class Ia molecules. Biology Direct 2006, 1:3doi:10.1186/1745-6150-1-3
Profiles in science
This is the biographies of some scientist. It is interesting. Have a look at it.
How to setup a default application for a file(like word with Microsoft word)
text files with gedit or nedit
pdf files with acroread or xpdf
xml files with gedit or nedit
For example
I want all "xml" file types to openwith gedit (text editor).
Right click the file and select "Open with" "other application" you will get window like this (shown below). If you want to choose different editor choose the file types available.

Select the file type you want. I have selected gedit (text editor)

Now i can open all the "xml"(any application) files with gedit text editor
May 19, 2008
How to submit sequence in genbank (NCBI)
 2)
2)
3)
 4)
4)
5)
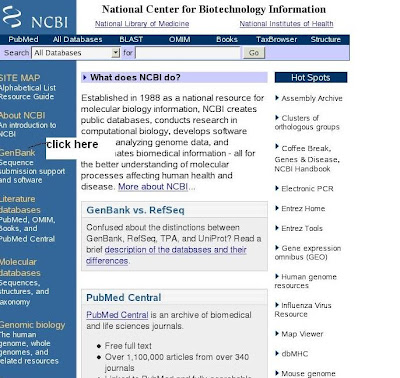
1) Open the NCBI home page (click genbank submission hyperlink)
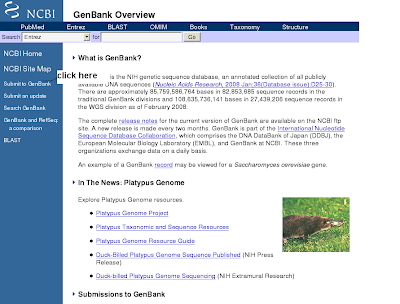
2) You will get genbank overview(click submit to genbank)
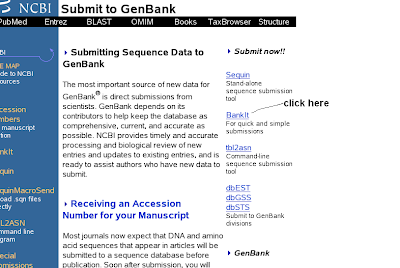
3) You will get submit to genbank (clikc the right side BankIt)
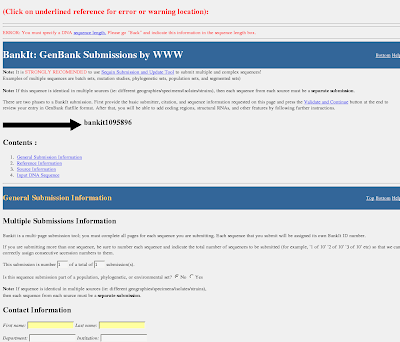
4) You will get BankIt submission form (enter sequence length)
5)You can fill the BankIt sequence submission form
May 15, 2008
INSTALl-postgresql in linux
This is a little more complicated than installing Apache.
The best bet is to install from packages since this will make things easier, both now and for upgrades.
If you decide to install from source code, you'll need to create a user and compile the source.
Create a PostgreSQL user (you'll need root access). This is who owns the system. I use the "postgres" user and group, you can use something different but it's best to keep it related to the package.
See your system documentation on how to do this (hint - man useradd). Give the user a home directory of /usr/local/pgsql as this is where PostgreSQL defaults to when it installs.
Once all that is done, untar the source:
$ tar -zxf postgresql-
or
$ tar -xjf postgresql-
Jump into the directory:
$ cd postgresql-
(replace
Again, configure has a lot of help:
$ ./configure --help
We'll just install into /usr/local/pgsql and leave the rest as-is:
$ ./configure --prefix=/usr/local/pgsql
Next we have to make it (this will take a while, it takes longer than Apache):
$ make
You will get a line:
All of PostgreSQL successfully made. Ready to install.
when everything is finished.
You'll need to log in as root to do the rest:
$ su -
Go back to the postgresql directory and do:
# make install
PostgreSQL doesn't allow root to start up the server for security reasons, so next you'll have to change the owner of the files:
# chown -R postgres:postgres /usr/local/pgsql
Change to the postgres user:
# su - postgres
Then, change to /usr/local/pgsql:
$ cd /usr/local/pgsql
We need to initialize a database cluster:
$ bin/initdb -D ./data
This will create a /usr/local/pgsql/data directory and initialize it ready for startup and use.
As usual, you can view options by using the --help switch:
$ bin/initdb --help
Then you can start:
$ bin/pg_ctl -D ./data -l data/logfile start
Now that postgresql is started, we need to create a database and another user before we go any further.
You should use a separate database for each of our projects, it's not essential though - it just makes things a little cleaner and easier to understand.
You should also use separate users for each database. This keeps everything separate and 'project a' won't be able to modify any of 'project b's data.
To create a new user in postgresql, it's pretty simple:
$ /usr/local/pgsql/bin/createuser
and follow the prompts - pretty simple :)
The new user should not be able to create new databases or create new users.
To create a database, it's a little different.
$ /usr/local/pgsql/bin/createdb --owner=username databasename
We have to set the owner of the database when we create it, otherwise the 'postgres' user owns it and then we have to grant access to allow our new user to access it.
Now that that's all done, log out of the postgres user, so we're back as root. In the postgres directory, the contrib/start-scripts/ directory has a few system start up scripts
Useful installation instruction from http://www.designmagick.com/article/2/Starting-Out/Installing-PostgreSQL
This is a little more complicated than installing Apache.
The best bet is to install from packages since this will make things easier, both now and for upgrades.
If you decide to install from source code, you'll need to create a user and compile the source.
Create a PostgreSQL user (you'll need root access). This is who owns the system. I use the "postgres" user and group, you can use something different but it's best to keep it related to the package.
See your system documentation on how to do this (hint - man useradd). Give the user a home directory of /usr/local/pgsql as this is where PostgreSQL defaults to when it installs.
Once all that is done, untar the source:
$ tar -zxf postgresql-
or
$ tar -xjf postgresql-
Jump into the directory:
$ cd postgresql-
(replace
Again, configure has a lot of help:
$ ./configure --help
We'll just install into /usr/local/pgsql and leave the rest as-is:
$ ./configure --prefix=/usr/local/pgsql
Next we have to make it (this will take a while, it takes longer than Apache):
$ make
You will get a line:
All of PostgreSQL successfully made. Ready to install.
when everything is finished.
You'll need to log in as root to do the rest:
$ su -
Go back to the postgresql directory and do:
# make install
PostgreSQL doesn't allow root to start up the server for security reasons, so next you'll have to change the owner of the files:
# chown -R postgres:postgres /usr/local/pgsql
Change to the postgres user:
# su - postgres
Then, change to /usr/local/pgsql:
$ cd /usr/local/pgsql
We need to initialize a database cluster:
$ bin/initdb -D ./data
This will create a /usr/local/pgsql/data directory and initialize it ready for startup and use.
As usual, you can view options by using the --help switch:
$ bin/initdb --help
Then you can start:
$ bin/pg_ctl -D ./data -l data/logfile start
Now that postgresql is started, we need to create a database and another user before we go any further.
You should use a separate database for each of our projects, it's not essential though - it just makes things a little cleaner and easier to understand.
You should also use separate users for each database. This keeps everything separate and 'project a' won't be able to modify any of 'project b's data.
To create a new user in postgresql, it's pretty simple:
$ /usr/local/pgsql/bin/createuser
and follow the prompts - pretty simple :)
The new user should not be able to create new databases or create new users.
To create a database, it's a little different.
$ /usr/local/pgsql/bin/createdb --owner=username databasename
We have to set the owner of the database when we create it, otherwise the 'postgres' user owns it and then we have to grant access to allow our new user to access it.
Now that that's all done, log out of the postgres user, so we're back as root. In the postgres directory, the contrib/start-scripts/ directory has a few system start up scripts
Useful installation instruction from http://www.designmagick.com/article/2/Starting-Out/Installing-PostgreSQL
May 13, 2008
NCBI Resource Locator
The NCBI Resource Locator provides stable, uniform addressing for NCBI content, making it easy to link to individual records. Some NCBI resources also provide services (like search) through these URLs.
How does it work?
Each URL has the form
http://view.ncbi.nlm.nih.gov///
Where:
* is an NCBI resource (e.g., pubmed, gene, nucleotide, etc.)
* is the action to perform (e.g., search, get, etc.). If is missing, the default verb "get" is used.
* is data used by the action to perform the request
Some examples:
* http://view.ncbi.nlm.nih.gov/pubmed/12345
Show the PubMed record with pmid 12345
* http://view.ncbi.nlm.nih.gov/pubmed/search/cancer
Search PubMed for "cancer"
* http://view.ncbi.nlm.nih.gov/gene/search/human+p53
Search PubMed for "human p53"
* http://view.ncbi.nlm.nih.gov/homologene/search/dystrophin
Search Homologene for "dystrophin"
Reference http://view.ncbi.nlm.nih.gov/
unzip file(bz2,tar) files
use the command "tar -cvf filename.tar" for unzip the tar files
use the command "tar -xzvf filename.tar.gz for unzip the tar.gz files
How to transfer files using SCP in linux
scp filename username@hostname(or ip address):/home/user/ (path)
for eg, in my homology machine i use
scp filename suresh@homology:/home/suresh/
May 12, 2008
What is the difference between hypothetical protein and putative protein
May 11, 2008
May 10, 2008
Michael Levitt's Thesis- online
Fold recognition server
This is one of the useful server for fold recognition. This has ten different servers implemented in it. It is useful server to select templates for modeling the protein molecule. https://genesilico.pl/meta2/
May 8, 2008
Mount pen drive in linux
" /sbin/fdisk -l " This will provide disks in your machine.
To mount pendrive - " mount /dev/sb2/ /mnt/cdrom/ "
socket error while fetching websites in scripting langauages
Site directed Insilico mutagenesis software
TRITON Home Page
May 7, 2008
Keyboard beep sound switch on/off
Go to Start Main menu - preferences-keyboard-preferences-sound-off.
Real passion - Dr. David Baker
Although he's only 42, David Baker is already a grandfather. Well...sort of.
Baker is raising a second generation of scientists in his lab at the University of Washington in Seattle. The Baker scientific family tree now includes scientists all across the country linked by a common goal: a driving curiosity to predict the shapes of proteins, the basic building blocks of our bodies.
Would you believe that all this happened in just 10 years?
Baker's remarkable enthusiasm for science and endless energy to solve hard problems keeps the family growing. This combination easily attracts new students to Baker's lab, and his captivating way with people keeps them there.
As with any good parent, Baker instills a sense ofindependence in his scientific children. After they leave the nest, most continue the journey in their own labs, where they raise their own research families.
Like glue, good communication holds everything together. Baker, a computational biologist, believes conversation gives birth to great ideas. Starting open discussions in the lab, he says, is one ofhis most important jobs.
"I remember a very lively energy in [David's] lab, "says Jeffrey Gray, who started working with Baker 5 years ago." David was the catalyst that increased the flow of ideas."
Gray, now a biomolecular engineer at Johns Hopkins University in Baltimore, Maryland, carries on the family tradition. Modeling his own career after Baker, Gray mentors many young scientists. Among them is a student who, as a high school senior under Gray's mentorship, placed fifth in the 2005 national Intel Science Talent Search competition (see sidebar, The Next Generation).
Birth of an Idea
Baker has spent much of his life tucked between two mountain ranges. A Seattle native, he hikes the local trails in the summertime and skis the slopes when the snow starts to fall.
"This is the greatest place on Earth!" Baker says. "The mountains are one of the great advantages of living here."
But for Baker, the mountains offer more than just pretty scenery and recreational opportunities. They symbolize an inner passion to achieve.
"David approaches science like he does a mountain," says Gray. "He finds the highest peak and heads toward it."
But what Baker now heads toward isn't what he originally set out to find. At first, he thought modeling the shapes of proteins was, well, boring.
"I remember writing a report for a college biochemistry course and thinking, 'Protein folding seems like a neat problem, but not much is happening in the field,'" Baker says.
He admits that his opinion changed during graduate school when he began studying how cells organize their many parts, which of course include proteins.
Our bodies consist of billions of proteins, large molecules made of smaller components called amino acids. Anywhere from a few to tens of thousands of amino acids link up in a particular sequence, and then each amino acid sequence folds into a unique three-dimensional structure.
It’s this shape that really determines a protein’s job. When a protein attaches to other molecules, it triggers a host of chemical reactions that run all of our biological machinery. “Proteins are incredibly organized and do amazing things,” says Baker.
But sometimes the things go wrong. Altering just one amino acid in the chain can change the entire shape of the protein. This switch can lead to life-threatening disorders like sickle cell disease or cystic fibrosis.
If we want to treat and prevent diseases, Baker says, we need to know what proteins look like. Having this information will help scientists custom-design medicines to target proteins and fix health problems.
The Shape of Things
At first glance, determining the structure of a protein from its amino acid sequence seems like it would be easy. But things have not turned out to be so simple. Score one for Mother Nature.
If a protein is really big, scientists can spend months or even years trying to determine its structure. Sometimes, knowing the shape of a similar protein and using that as a guide can speed up the process. But part of the problem is that researchers only know the structures of a small fraction of the proteins in the human body.
So where does that leave scientists who want to find out a protein’s shape? They either do physics-based experiments with X rays or huge magnets, or they use computer models to make good guesses.
Both approaches have their drawbacks. X-ray crystallography and NMR spectroscopy, the methods from physics, are labor-intensive and can be expensive. The computer modeling approach can be inaccurate and unreliable.
For Baker, the Holy Grail is developing software programs that generate high-resolution models of proteins. Ideally, these models would reveal every feature of a protein's landscape, including its atoms, hydrogen and other bonds, and all the places where important chemical reactions occur.
With refined pictures, researchers can examine single proteins and track their interactions with other molecules. Accurate models could ultimately let researchers make entirely new proteins with custom functions, motions, and chemical reactions.
Scientists have been trying to develop accurate computer models for years. But the models rarely capture all the details, instead creating mostly “rough sketches” of how protein parts fold together into complex structures.
“Simplifying the model of a protein is like smoothing out a mountain until you have rolling hills instead of sharp peaks and deep valleys,” explains Baker, adding that a lot of extra work goes into finessing the computer’s output.
Quality Time
With an intense interest in trying to solve what others find unsolvable, Baker splits his time between his two families— his wife and two children and his lab personnel. During the week, he spends regular work hours with about a dozen postdoctoral fellows, 10 graduate students, and a handful of other researchers. Many come from different countries and different scientific fields, like chemistry, engineering, and medicine.
“It’s a privilege to walk out of my office and talk to really smart people interested in the same problem I am,” he says. “I feel very lucky to be here.”
Together, the group focuses on a computer software program that Baker developed called Rosetta. Just like the famous stone of the same name once helped linguists decipher ancient languages, Baker and his group hope their Rosetta will decode the mysterious shapes of proteins and even help them build new and better versions.
Basically, Rosetta uses information about a protein’s amino acid sequence to predict its possible shape. It breaks the protein into small chunks of amino acid sequences, searches for all the different shapes each chunk could assume, and then mixes and matches them until it finds a perfect fit. Rosetta may create up to 10,000 simulations and run for 100 days before honing in on the structure of even the simplest protein.
Baker and his team have created many Rosetta flavors, each of which can answer a different question, such as how a protein interacts with another protein or with a DNA sequence. Some varieties incorporate experimental data or the structural information of other, similar proteins.
One version of Rosetta being developed could run on the computers in University of Washington dorm rooms when students aren’t using them. This could add up to 10,000 processors to the team’s protein structure prediction effort and make the work go faster.
Because Baker wants as many minds as possible working on the problem, he gives Rosetta to other scientists for free.
Baker and his students take the Rosetta models and go to work refining them. Sometimes, they run into problems, but that doesn’t stop progress. When this happens, Baker says he knows it’s time to talk, and he brings the lab together to troubleshoot.
“I think the human factor is one of the most important elements [of science],” Baker says.
Outward Bound
Whether the lab takes the day off to go hiking or sits around the lunch table trying to solve a problem, Gray says, “David has the energy to push people beyond their boundaries to explore new ideas.”
Baker’s ultimate goal is predicting a detailed structure at a level of resolution, or clarity, of 2 angstroms, or 200 billionths of a millimeter.
“If we can successfully model protein structures with a level of accuracy so that biologists are confident the models are right, we could compute all the protein structures that already exist without [doing] experiments,” Baker says.
This, he notes, would save researchers a lot of time and money.
Some people may find it ironic that Baker, who never took a computer course in his life, not only developed a tremendously useful software program but also spends his days (and sometimes nights) thinking about computers.
But he doesn’t see it that way.
Baker says he finds a problem he wants to solve, meets up with a great group of people, and then learns whatever he needs to know along the way.
Every year, Baker recruits more adventurers to join him on his climb to model proteins. Only together, he says, can they reach the top—better models of protein structures that may lead to new drugs and vaccines for keeping us healthy.
New Direction
One of the latest Baker family franchises involves designing new proteins not found in nature. Unlike the usual approach of starting with an amino acid sequence and then building a structure, Baker and his team are working backwards. They’re designing proteins from scratch.
Like architects who design a house before drawing up blueprints, Baker and his trainees began with a sketch of a made-up protein structure. Using Rosetta, they pieced together a string of amino acids that most likely would link up to create the new protein, and then made the actual protein in the lab. In an early experiment, the researchers found that the real protein was virtually identical to the one Baker had imagined.
The scientific community recognized this work as monumental and Baker and his research group received a prestigious prize for the best paper published in a 2003 issue of the journal Science.
Next, Baker wants to design proteins that cause particular chemical reactions on demand.
“This would open up a whole new world of functional proteins,” says Baker.
The ability to create proteins made to order offers a promising route for developing custom proteins that could interrupt or enhance a particular reaction inside a cell.
Community Center
When it comes to modeling protein structures, Baker and his group have proven that they can climb with the best. Every other year, the group enters a friendly competition called CASP (Critical Assessment of Techniques for Protein Structure Prediction). They go head to head with hundreds of labs worldwide to see who can make the best predictions.
In December 2004, scientists from more than 200 labs gathered in Italy, submitting a total of 15,000 predictions for selected protein structures. The only people who knew what the proteins actually looked like were the judges.
Baker’s group used Rosetta to develop their models, and as in previous years, Baker’s team did very well. One of the postdoctoral researchers in Baker’s lab modeled a protein structure with a very small average error of 1.59 angstroms.
“I like working on the methods and seeing them pay off,” says Baker. “CASP is very collegial and a great experience for the people making the predictions.”
Although there are some competitive aspects, everyone walks away from CASP with a prize—the opportunity to work together, learn about current challenges, set future goals, and assess the methods and technology used to predict protein structures. For this reason, scientists prefer to call CASP a “community-wide experiment” instead of a contest.
Despite this progress, and even with Baker’s many successes, a lot still needs to be done. When it comes to accuracy, many current, low-resolution models are in the ballpark, Baker says, but they have a way to go.
As Baker and his team continue to work on the problem, one thing stands in their way: insufficient computing power.
“For a long time, the problem was not having accurate descriptions of proteins and their interactions,” explains Baker. “But now the problem is that we don’t have enough computer power to run the simulations.”
For example, Rosetta can run for months before it finally spits out a model that closely resembles the real thing. Not only does this take computer time, it also takes a lot of computing power, Baker says.
Making really accurate predictions, and lots of them, means having a herd of computers that can quickly process data. Currently, Baker is talking to large computer companies to try to get his hands on more machines, especially ones with faster processors.
Family Reunion
Every summer, Baker invites his extended scientific family to join him in Seattle for what he calls “Rosetta Commons.” For 2 days, they talk about prediction projects, challenges they’re encountering, and potential ways to improve the software behind it all.
“We’ve all started labs that are working on different problems,” says Gray, who attended the reunion last summer. “But we’re still related by the Rosetta code.”
On the third day, the group usually heads for the hills, something the former students fondly remember from their days in Baker’s lab.
“If you’re walking next to David, you’re going to be talking about science,” jokes Gray. “David focuses so much on science. It’s what he does naturally.”
Last summer, the group hiked up Dragontail Peak, which looms about 9,000 feet above sea level. The trail, recalls Gray, was quite ambitious.
Minus the time for picnicking and swimming in a crystal-blue lake, the group spent nearly the entire day climbing to the top. They were only halfway down when the sun started to set.
“It was 8:00 p.m., and we still had several hours of hiking,” says Gray.
Without enough flashlights to guide their way down, Baker and former postdoctoral researcher Brian Kuhlman—by far the most experienced hikers in the group—volunteered to run back to the cars, drive into town, and bring back extra supplies. The two met up with the other hikers, still creeping their way down, with flashlights and chocolate.
Everyone finished, still in good spirits, remembers Gray.
“It was definitely a bonding experience!” he says.
May 3, 2008
Non-scientists think research is not a 'real job'
Non-scientists think research is not a 'real job' because scientists don't have deadlines or discrete targets. On the contrary, I say, we are accountable to those who fund us. Investors want financial returns, taxpayers demand medical advances. We are judged by the number and impact factor of our publications. I have friends who have actually calculated the average impact factor of papers published at their institution, hoping to gauge their own competitiveness.
At the Biopolis, we are subject to annual evaluations in which our productivity is rated on a scale of 1 to 5. These determine our bonus and possibly our career prospects. You don't get points for effort: my decision to study a novel protein has proved unwise because I cannot use established reagents and protocols to churn out data and papers. I can ask many questions about a protein of unknown function. But as I was specifically instructed to "focus on publishing my work as it is completed", the key question driving my research must be: what is the minimum amount of data that can coalesce into a paper?
This practical approach is difficult to reconcile with the risk associated with novel or creative projects. It is, however, necessary if I want to stay employed in a world dictated by 'key performance indicators'. We are subject to constant selection pressure. And, to paraphrase Darwin, those who can adapt, survive.
from Nature Jobs section in Nature May 5th issue
Reference manager-tool required for the scientific community
Programs promise to end PDF paper-chase. Researchers are buzzing about a new type of software that allows them to manage their research paper downloads from online journals much more effectively. http://www.nature.com/news/2008/080430//full/453012b.html
Required skills for a computational biologist
Python programming language.
Competent in perl programming language
Experience of shell scripting
Working in a Unix / Linux environment.
A good working knowledge of sequence analysis
Experience in functional annotation of biological sequences
SQL/PostgreSQL database design and use
Statistical analysis
Good knowledge of online Bioinformatics resources
HTML, CSS, PHP etc.
An advice to Indian bioinformaticians (so called)
Applying for a position in my group
On average twice per year I hire a PhD student or a post-doc to work with us on one of the two main topics of the group. This stands in a marked contrast to the average of two people per day who ask if I have a job for them. So, what should you do to make more than the 0.5% chance that you have by luck alone?
First of all, I am not different from any other group leader. I only look at applications from people who show immediately in the first three lines of their letter that they have actually spent time preparing the application. Applications from people who only present themselves and just ask for any job are not taken serious. Applications that start with "Dear sir" will not be read. It is YOUR duty as applicant to find out what my group is working on, what kind of people we are looking for, and what you can mean for the group. To do so, look at our WWW page and look at the articles written by the group members over the last three years. If that gives you the impression that you could add something to the group, then you should think how to formulate that concisely and comprehesively without looking very dumb or very arrogant. Make sure the things I must know are on page 1 of the application. Useless information like hobbies, the family name of your mother, the fact that you went to some high-school somewhere, the fact that you have a drivers license, etc., should be skipped, or at best placed on the last page. Make sure that you explain which of your previous experiences you think are relevant for working in our group.
If you work outside 'the west', you have an extra handicap. Not that we simply assume that people from south-america, africa, or south-asia are by definition dumb, but it is very difficult for us to judge what your papers and courses are worth. And flying you in for an interview is too expensive. Coming from outside 'the west' your best bet is a letter of recommendation from a professor with extensive experience in 'the west'. Obviously, a few articles in international journals with you as first author will help very much too. If you write your master-thesis, write it in English and put in somewhere on the WWW so we can see 'how good you are'. There are several other trivial tips. But in general, make sure that we get a good opportunity to judge you, because that is your major bottleneck, being properly evaluated.
Make sure you understand how science is organized in the country where you want to go to. For example, I often get letters from people from India and Pakistan who want to do an internship. As I don't have any idea what that means, I normally answer 'no, sorry'. Obviously, to you it seems stupid that I don't know what an internship is, but my country organized science differently. And as it is you who wants something from me, and not the other way around, you should make sure that I know what you want and what you have to offer.
The points listed above are very general and hold for each job application. If you want to explicitly work with me, you should be a smart scientist, but also a good programmer. We design software to solve biomedical problems. And we will not spend time teaching people how to write software. You should be able to start programming on day 1. I therefore want to see the source code of your last programme of 10000 lines or more. No need to apply when you cannot provide that. (It doesn't need to be scientific software, a game, or a programme to administrate the inventory of the student club is fine too; just as long as it is your software).
Obviously, you can try to write an open application, but that only makes sense if you are a very exceptionally brilliant person, the kind of person so much worth having that we just raise the funds to hire you. Otherwise, look at the list of open position below and see if your job is listed there, and if so apply for the job, using the hints given above.
Good LuckGert Vriend
http://www2.cmbi.ru.nl/groups/modeling-and-data-mining/jobs/
May 2, 2008
Scripting the Web with PHP
PHP is one of the most popular open source scripting languages for the Web. It’s fast, effective, and very easy to learn. See for yourself. http://www.linux-mag.com/id/820
Creating PDFs in Linux
Sharing documents such as papers, reports, and specifications is made easy with Adobe’s Portable Document Format (PDF). Here’s a primer on how to create PDFs on Linux.
http://www.linux-mag.com/id/1983
May 1, 2008
Chimera - Molecular Modeling software
So, now that you are convinced to give Chimera a try, how do we make some movies? There are tutorials on the web site, but to start with, you can try this simple set of commands. In your favorite text editor (Notepad in Windows or TextEdit in Mac OS X), create a file called “movie.cmd” and enter the following text:
movie record
roll y 1 360
wait 360
movie stop
movie encode
Save the file, start Chimera and load your molecule of interest. Get it looking how you like, then open the file “movie.cmd” (File… Open…) and watch it go. This script will rotate the molecule about the y axis in 360 steps of 1 degree, then save the movie. When it’s done you should have a file named “chimera_movie.mov” that you can show to your students. If you would like to see some of the movies I use in my Genetics and Biochemistry lectures, or if you would like to use them in your own classes, please visit my website.
- E.F. Pettersen, T.D. Goddard, C.C. Huang, G.S. Couch, D.M. Greenblatt, E.C. Meng, and T.E. Ferrin (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 25(13): 1605-12.
Reference : Education corner , spring 2008, RCSB Newsletter, Number 37.
Online Bookmark
What is del.icio.us?
del.icio.us is a collection of favorites - yours and everyone else's. You can use del.icio.us to:
- Keep links to your favorite articles, blogs, music, reviews, recipes, and more, and access them from any computer on the web.
- Share favorites with friends, family, coworkers, and the del.icio.us community.
- Discover new things. Everything on del.icio.us is someone's favorite -- they've already done the work of finding it. So del.icio.us is full of bookmarks about technology, entertainment, useful information, and more. Explore and enjoy.
del.icio.us is a social bookmarking website -- the primary use of del.icio.us is to store your bookmarks online, which allows you to access the same bookmarks from any computer and add bookmarks from anywhere, too. On del.icio.us, you can use tags to organize and remember your bookmarks, which is a much more flexible system than folders.
You can also use del.icio.us to see the interesting links that your friends and other people bookmark, and share links with them in return. You can even browse and search del.icio.us to discover the cool and useful bookmarks that everyone else has saved -- which is made easy with tags.
Biobar tool bar in firefox
- Biobar
- A power browsing toolbar for the Life Sciences (Biology/Bioinformatics/Medicine). Easily search and retrieve data from over 45 bioinformatics services from the comfort of the toolbar. Links to bioinformatics tools and resources. You can add/replace/change the searched databases and associated terms by following instructions given at http://biobar.mozdev.org/Customize.html.
Apr 19, 2008
PDB crossed more than 50,000 structures
With this week's update, the PDB archive reached a significant milestone in its 37-year history: The holdings now contain more than 50,000 current experimental structures.
Genome Sequenced
http://www.genomesonline.org/
Protein-Protein docking server
UCSF Chimera
This is very useful tool for graphical visualization. Taking pictures using chimera is the best. Even compared to commercially available molecular modeling softwares. check it out this http://www.cgl.ucsf.edu/chimera/
Apr 18, 2008
India Takes an Open Source Approach to Drug Discovery
Volume 133, Issue 2, 18 April 2008, Pages 201-203 , Cell
Apr 17, 2008
Getting Started in Text Mining
http://www.ploscompbiol.org/article/info%3Adoi%2F10.1371%2Fjournal.pcbi.0040020
Good English dictionary site
Indian journals push for clinical-trial registration
Eleven of India's leading medical journals will now consider publishing the results of clinical trials only if the trial in question has been registered with the Indian Council of Medical Research (ICMR) in New Delhi or any primary clinical-trial register.
The move follows the 2005 decision by the International Committee of Medical Journal Editors to publish only the results of registered trials. According to Kanikaram Satyanarayana, deputy chief of the ICMR, the delay was partly due to reluctance by the journal editors and pressure from industry. In addition, the ICMR had hoped that a bill it drafted in 2006 to regulate human trials would become law.
Trials that start in or after June of this year must be registered before enrolling their first participant; those beginning before this must register retrospectively. The ICMR says mandatory registration will ensure transparency and honesty, and discourage unethical trials.
An estimated 250 drug trials are under way in India, with applications for 30 more received on average each month.
Online file format conversion
http://www.zamzar.com/
List of published biological databases
Useful one.
Protein-Protein Interaction through text mining tool
Biodegradation Server(BPD server)
Interesting article in Biodegradation
The environmental fate of organic pollutants through the global microbial metabolism
The production of new chemicals for industrial or therapeutic applications exceeds our ability to generate experimental data on their biological fate once they are released into the environment. Typically, mixtures of organic pollutants are freed into a variety of sites inhabited by diverse microorganisms, which structure complex multispecies metabolic networks. A machine learning approach has been instrumental to expose a correlation between the frequency of 149 atomic triads (chemotopes) common in organo-chemical compounds and the global capacity of microorganisms to metabolise them. Depending on the type of environmental fate defined, the system can correctly predict the biodegradative outcome for 73–87% of compounds. This system is available to the community as a web server (http://www.pdg.cnb.uam.es/BDPSERVER). The application of this predictive tool to chemical species released into the environment provides an early instrument for tentatively classifying the compounds as biodegradable or recalcitrant. Automated surveys of lists of industrial chemicals currently employed in large quantities revealed that herbicides are the group of functional molecules more difficult to recycle into the biosphere through the inclusive microbial metabolism.
Apr 15, 2008
Molecular dynamics paper in science
Arkin IT, Xu H, Jensen MØ, Arbely E, Bennett ER, Bowers KJ, Chow E, Dror RO, Eastwood MP, Flitman-Tene R, Gregersen BA, Klepeis JL, Kolossváry I, Shan Y, Shaw DE.
D. E. Shaw Research, New York, NY 10036, USA.
Science. 2007 Aug 10;317(5839):799-803.
Na+/H+ antiporters are central to cellular salt and pH homeostasis. The structure of Escherichia coli NhaA was recently determined, but its mechanisms of transport and pH regulation remain elusive. We performed molecular dynamics simulations of NhaA that, with existing experimental data, enabled us to propose an atomically detailed model of antiporter function. Three conserved aspartates are key to our proposed mechanism: Asp164 (D164) is the Na+-binding site, D163 controls the alternating accessibility of this binding site to the cytoplasm or periplasm, and D133 is crucial for pH regulation. Consistent with experimental stoichiometry, two protons are required to transport a single Na+ ion: D163 protonates to reveal the Na+-binding site to the periplasm, and subsequent protonation of D164 releases Na+. Additional mutagenesis experiments further validated the model.
Apr 14, 2008
Secrets of Greatness
Progress toward Public Access to Science
http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371/journal.pbio.0060101&ct=1
Apr 13, 2008
Notes to a young computational biologist
Want to be a UN Volunteer Online?
Steps to reach my goal
computer help
http://www.computerhope.com/
Apr 12, 2008
Interview with DG-CSIR
Read this interview with Director general. Especially the first anonymous comment.
http://spicyipindia.blogspot
Its about my coimbatore
http://en.wikipedia.org/wiki/Coimbatore
Briefings in Bioinformatics first impact factor is 24
http://bib.oxfordjournals.org/cgi/content/full/8/4/207
A Nobel Warrior
http://www.time.com/time/magazine/article/0,9171,1670524,00.html
Ten simple rules
http://www.ploscompbiol.org/search/simpleSearch.action?query=ten+simple+rules&x=0&y=0
On the Process of Becoming a Great Scientist
A article in plos computational biology by Morgan C. Giddings
http://www.ploscompbiol.org/article/info:doi%2F10.1371%2Fjournal.pcbi.0040033
Nobel prizewinner's paper retracted
http://www.nature.com/news/2008/080305/full/452013a.html
Water Muddy waters
India's population is growing, and its water supplies are not keeping pace. Can an ambitious scheme to connect up the country's rivers slake the nation's deepening thirst? Daemon Fairless investigates.
Malaria -eradication
Malaria Eradication in India: A Failure?
In the 7 December 2007 issue, L. Roberts and M. Enserink discuss malaria eradication in the News Focus story "Did they really say … eradication?"http://www.sciencemag.org/cgi/content/full/319/5870/1616d
Biotech in Bangalore
http://www.nature.com/naturejobs/2008/080403/full/nj7187-660a.html

.jpg)








